About usage
RVFscan provides a multi-annotation system for identifying the important VFGs of 24 pathogenic
bacteria (see “Species list” in Homepage). This analysis tool can be applied directly to reads
for VFG prediction without assembly, and especially suitable for clinical metagenomic data with
low-biomass and high complexity.
Here, the usage serves as a reference for how to add an analysis task for RVFscan and interpret
analysis results.
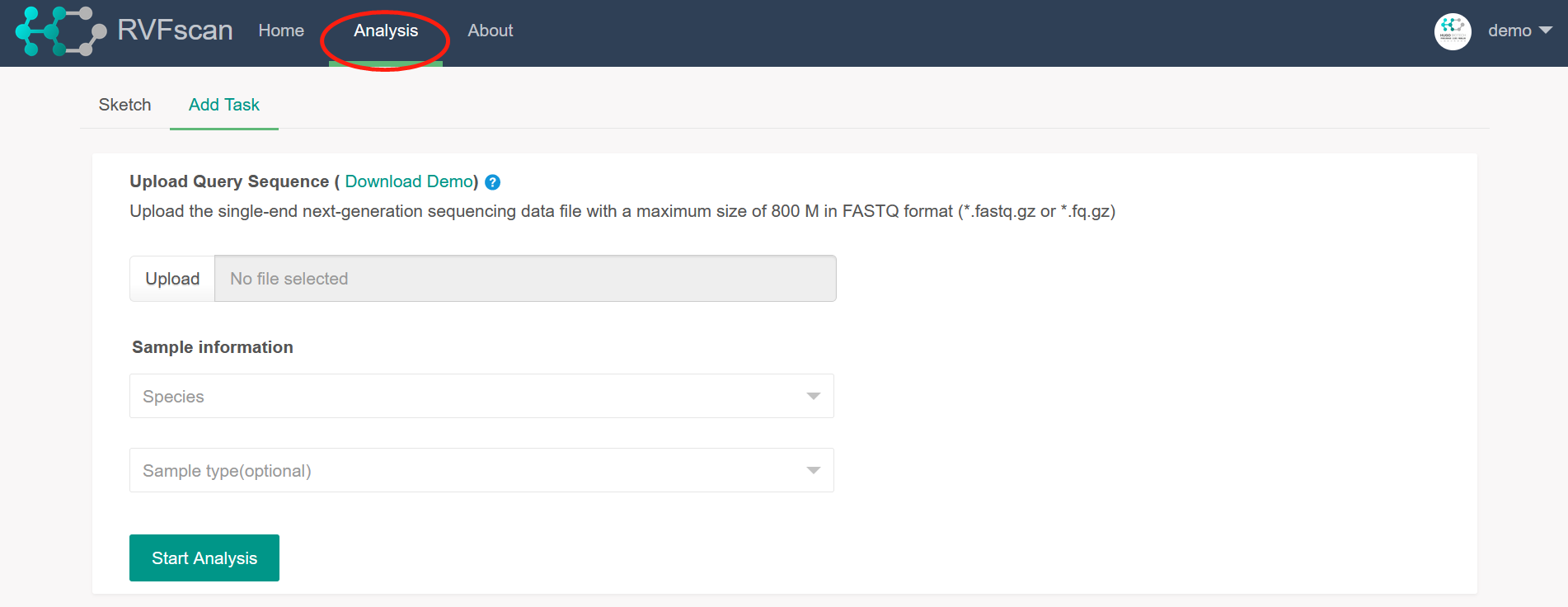
Add Task
Users can perform the analysis by the single-end next-generation sequencing data, especially for
the clinical metagenomic sequencing data with Illumina platforms.

-
Select and upload the local query sequence file by clicking on the ‘Upload’ button. This
requires the file with a maximum size of 800 M in FASTQ format (*.fq.gz or *.fastq.gz). And
only one file is uploaded at a time.
-
Select the species information for the query sequence data from the drop-down menu of 24
detectable species. This tool does not accept VFGs identification outside the 24 species. If
the
data involves multiple species, user can select more than one species at the same time. If
the
species involved in the data is uncertain, user can select ‘Unknown’. Please note that only
VFGs
of the species selected here are shown in the analysis results.
-
Select the sample type for the query sequence data from the drop-down menu of 6 sample
types. For sample that do not belong to the first five types, user can select ‘Others’. This
term is optional.
-
Click the ‘Start Analysis’ button to submit background analysis.
Here, a demo file of the demo sequence data serves as a example for acceptable sequence data.
User can download the demo file and perform analysis by uploading it and filling in the sample
information. This demo sequence data involves four species, including
Acinetobacter baumannii,
Klebsiella pneumoniae,
Staphylococcus
aureus and
Streptococcus pneumoniae.
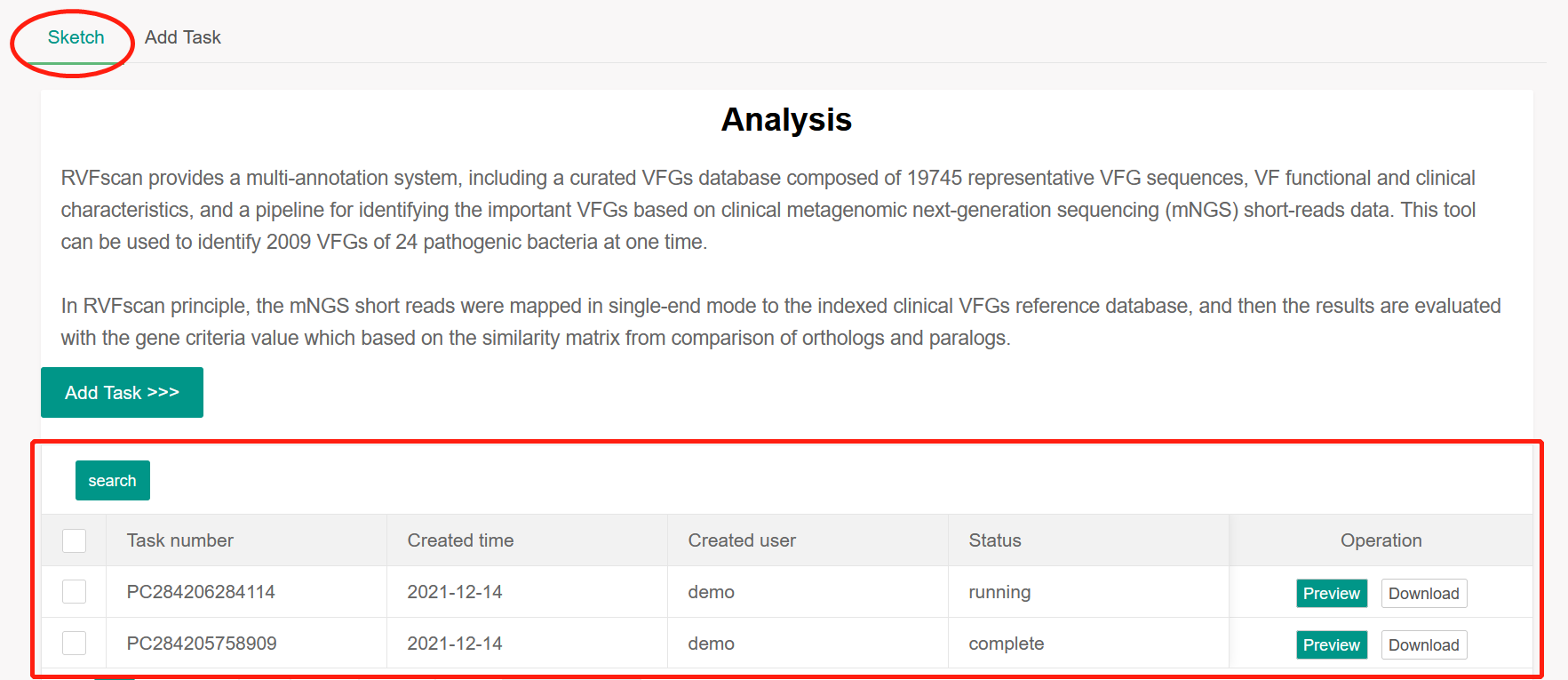
After the task is added successfully, the task list page is displayed. If the task status is
‘Standby’ or ‘Running’, the task is queuing up or being performed in the background,
respectively. When the analysis is completed, user will receive a reminder email from which can
link to the task list page. At this point, the task status is ‘complete’, and user can click the
‘Preview’ button to preview the results. For user convenience, the task list supports searching
by task number and created time.
Interpret results
Enter the interface of preview results by clicking the ‘Preview’ button in the task list. This
page displays the species selected by the user and the results list. The list includes
identified VFG symbol, the count of uniquely mapped reads, species, VF name, VF function, VFG
function (GO, EggNOG).
NOTE: Analysis results were reported for all VFGs supported with at least one unique read,
without manual filtering. For species with a low count of mapped reads, the VFG results need to
be carefully considered.
Download results
For the interface of preview results, user can download the result list file and the
intermediate result file, respectively, by clicking the download icon in the upper right corner.
Users can download TXT or XLS files.
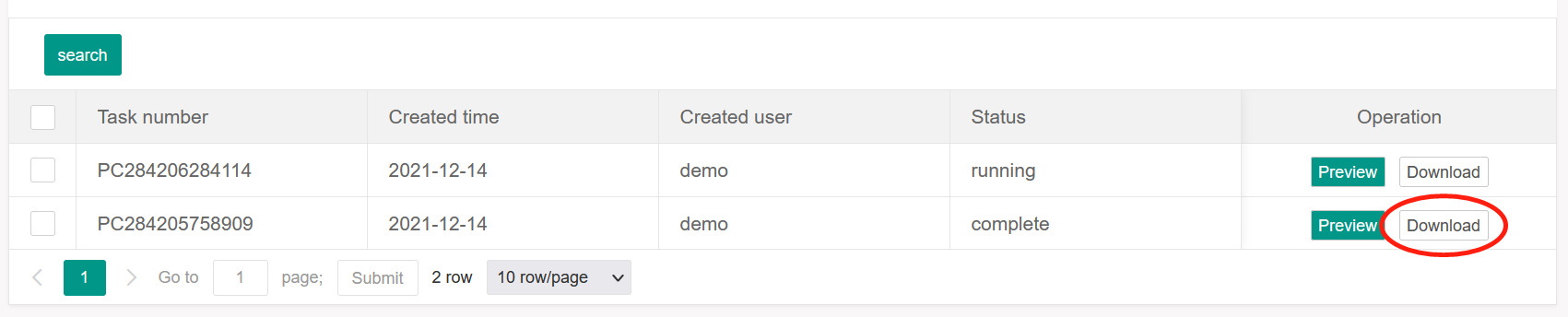
For the task list, user can download the zip file, including the result list file and the
intermediate result file, by clicking the ‘download’ button in the Operation column.
NOTE: The tasks information in task list and analysis results are reserved for 7 days. Please
save and download the required files before becoming invalid.
In addition, in the spirit of academic collaboration, we provide the detection rates and HF VFG
profiles of 24 pathogens for academic use. To obtain permission, you will send your name,
organization, purpose of research to rvfscan@hugobiotech.com via your institutional email
address.
The annotation of VFGs function was refer to The Gene Ontology knowledgebase (GO,
http://geneontology.org/), UniProt (https://www.uniprot.org/uniprot/) and EggNOG
(http://eggnog5.embl.de/#/app/home, v5.0).
Data and data products from GO contributed after 1999 and prior to 2019 (release date
"2021-09-10" and "doi: 10.5281/zenodo.4033054") are licensed by Gene Ontology Consortium under
the Creative Commons Attribution 4.0 Unported License unless otherwise marked
(https://creativecommons.org/licenses/by/4.0/).
For data and data products from UniProt, they have chosen to apply the Creative Commons
Attribution (CC BY 4.0) License (https://creativecommons.org/licenses/by/4.0/) to all
copyrightable parts of the databases.